Product Specification
Store at 2-8oC away from the light.
Materials
Reagents required:
• GoldenSelect DNA Size Select Beads
• 70% ethanol freshly prepared from 200 proof non-denatured ethanol
• Nuclease free water or 10mM Tris-Acetate pH8.0
Consumables and Hardware required:
• Automation compatible 96-well reaction plates (4titude FrameStar 96 semi-skirted.
PN:4ti-0770/C), or 4titude 8-strip PCR tubes (PN:4ti-0792) or 1.5ml tubes
• 4titude foil seals (PN:4ti-0536) or 4titude 8-strip caps (PN:4ti-0751)
• Magnetic separation devices. For 96 well plates: ALPAQUA Magnum FLX Enhanced
Universal Magnet Plate (SKU:A000400). For 0.2ml 8-strip PCR tubes and 1.5ml tubes:
Permagen Magnetic Separation Rack (PN:MSR1224B)
• Liquid Handling Instruments for Automation or multi-channel hand pipette
General Procedure for DNA fragment size selection
Please see Figure 1 Round 1 size selection, lane B1 to A2, to use as a guide to
select appropriate ratios of the beads to use to recover specific size range of
DNA fragments.
1. Bring the DNA Size Select Beads to room temperature and mix thoroughly by gentle
shaking.
2. Add desired ratio/volume of the DNA size select, i.e. 18μl of the beads to 10μl DNA
solution.
3. Mix the DNA Size Select beads and DNA solution thoroughly by vortexing for a few
seconds.
4. Incubate at room temperature for 10 minutes or mix for 10 minutes on a roller mixer.
5. Centrifuge the mixture briefly and place the tube on a magnetic separation rack to
separate beads with bound DNA from solution for 5 minutes or until solution becomes
clear. Larger sample volumes may require more time for separation.
6. Retain the tube on magnet and aspirate the cleared solution from the tube and discard.
Do not disturb beads while removing supernatant.
7. Dispense 200μl of 70% ethanol to each sample and incubate for 30-60 seconds at room
temperature. Remove the ethanol and discard. Repeat for a total of two washes. Samples
must remain on magnet during these washes.
8. Place the reaction plate on benchtop and air dry about 10 minutes. Do not over dry the
beads as this will reduce elution efficiency.
9. Add desired volume of elution buffer of your choice (10mM Tris-Acetate pH8.0,
nuclease free H2O, etc.) to each sample and vortex 30 seconds. Centrifuge briefly and
incubate the tube at 60C for 10 minutes or at room temperature for overnight for
maximal recovery of DNA fragments from the beads.
10. Place samples on magnet for 3 minutes or until solution is clear and transfer supernatant
to a fresh tube.
Applications
Single Size Selection of DNA fragments and recovery of unbound fragments example:
Note: Size of DNA as well as concentration and volume of the sample will affect the yield.
Table 1. Round 1 size selection: Lower ratio will select for larger fragments. Round 2 will recover all fragments that did not bind to the beads during round 1 size selection.
Round 1 size selection:
Input DNA = 20μl of GeneRuler 100bp DNA ladder diluted 1:4 in nuclease free water.
Step 1. Add desired ratio/volume of the DNA size select, i.e. if the ratio of 1.0X is desired,
then add 20μl of the beads to 20μl DNA solution. Ratio used in this example: 0.5X – 2.0X.
Follow the steps 2-5 of General Procedure for DNA fragment size selection.
Step 6. While retaining the tubes on magnet, removed the cleared solution from the reaction
tubes and transfer to fresh tubes and save for recovery of unbound DNA fragments. Do
not disturb beads while removing supernatant.
Follow step 7-10 for washing and elution of the bound DNA fragment. DNA was eluted in
20μl of nuclease free water.
Round 2-recovery of unbound DNA fragments from Round 1 size selection.
Add a fresh aliquot of the DNA Size Select Beads to the supernatant saved from step 6 of
Round 1 size selection (see Table 1 above) at combined total ratio = 2.5X starting sample
volume.
Follow General Procedure for DNA fragment size selection for binding, washing, and
elution. DNA was eluted in 20μl of nuclease free water.
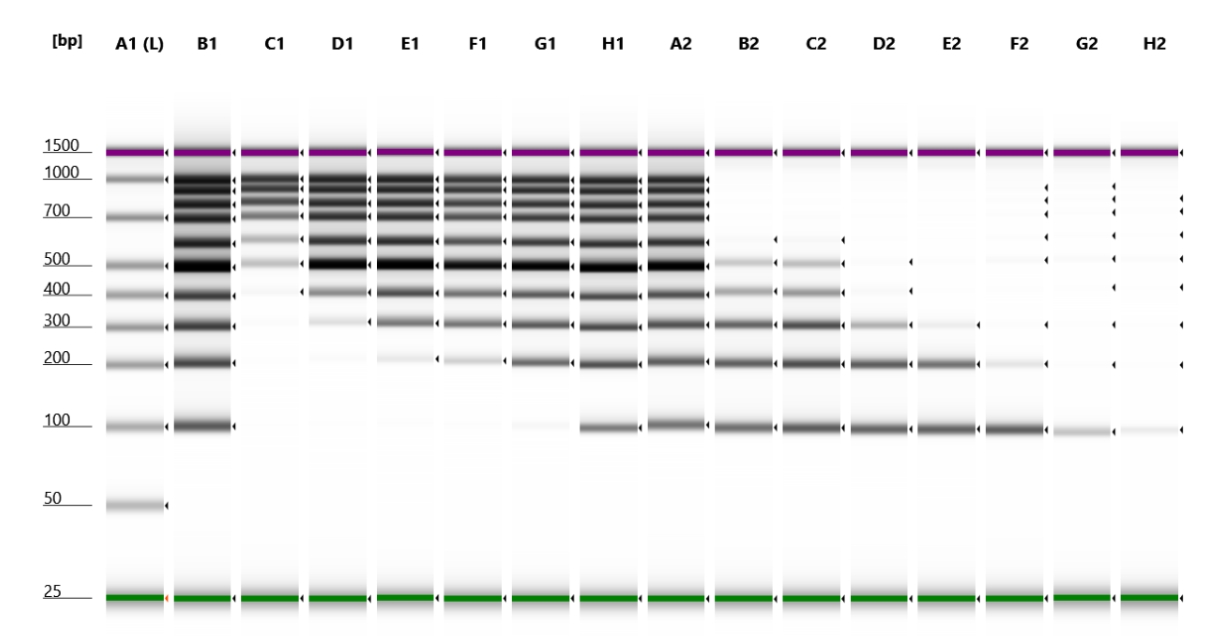
Fig. 1 TapeStation D1000 Tape data of Single Size Selection.
A1: Ladder
B1: Input GeneRuler 100bp ladder
C1: Round 1 – 0.5X
D1: Round 1 – 0.6X
E1: Round 1 – 0.7X
F1: Round 1 – 0.8X
G1: Round 1 – 1.0X
H1: Round 1 – 1.5X
A2: Round 1 – 2.0X
B2: Round 2 – 2.0X
C2: Round 2 – 1.9X
D2: Round 2 – 1.8X
E2: Round 2 – 1.7X
F2: Round 2 – 1.5X
G2: Round 2 – 1.0X
H2: Round 2 – 0.5X

